Evaluation of predicted data
To convert the overlap between predicted bounding boxes and ground truth bounding boxes into a measure of accuracy and precision, the most common approach is to compare the overlap using the Intersection over Union (IoU) metric. IoU is the ratio of the overlap area of the predicted polygon box with the ground truth polygon box divided by the area of the combined bounding box.
The IoU metric ranges from 0 which is not overlapping at all to 1 which is totally overlapping. In the wider computer vision literature, the common overlap threshold is 0.5, but this value is arbitrary and ultimately irrelevant to any particular ecological problem. We treat boxes with an IoU score greater than 0.4 as true positives, and boxes with scores less than 0.4 as false negatives. A value of 0.4 was chosen for threshold-based visual assessment, which indicates good visual agreement between predicted and observed crowns. We tested a range of overlap thresholds from 0.3 (less overlap between matching crowns) to 0.6 (more overlap between matching crowns) and found that 0.4 balances the strict limit without erroneously deleting for downstream use Analyzed trees.
For this blog, we will be using NeonTreeEvaluation dataset which will be attached at the bottom of the blog.
Here we calculated the evaluation score by resampling the image with various resolutions and then judging model predictions on those various resolutions and we did it because overall the models are too sensitive to input resolution. There are two strategies to improve, the preprocessing, and the model weights so what we thought of we can retrain the model using different zoom augmentations to try to make it more robust to input patch size.
So first step was to get evaluation data and then resample the image on various resolutions
where image_path is the path to the image which is the path to the original image and resample_image is the path to the image which has been wrapped in a certain resolution and new_res is the resolution on which the original image would be wrapped.
Now we calculated the evaluation score for the resampled image
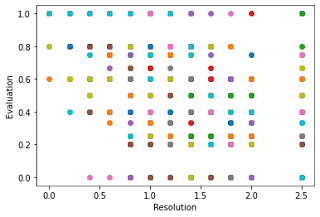
First, we resampled the image and then predicted the trees on that particular wrapped image and got the CSV file for the predicted data, and after that, we evaluated the results given by our model by our deepforest function model.evaluate and the recall is the proportion of ground truth which have a true positive match with a prediction based on the intersection-over-union threshold, this threshold is default 0.4 and can be changed in model.evaluate(iou_threshold=<>), and we gonna plot resolution on x axis and evaluation score on y axis and plotting can be done by the following snippet
and snippet to execute the following code is
where resolution_values has values on which image would be wrapped that is if 0.20 means 20cm, the image gets so blurry on resolution above 100cm so it doesn't make any sense to evaluate these images but we still evaluated to check what the model predicts on those resoluted images.
We used 1000 files on this for the evaluation and plotted the results or it
Colab notebook: code
Comments
Post a Comment